Peter Antal
Circuits, Features, and Heuristics in Molecular Transformers
Dec 10, 2025Abstract:Transformers generate valid and diverse chemical structures, but little is known about the mechanisms that enable these models to capture the rules of molecular representation. We present a mechanistic analysis of autoregressive transformers trained on drug-like small molecules to reveal the computational structure underlying their capabilities across multiple levels of abstraction. We identify computational patterns consistent with low-level syntactic parsing and more abstract chemical validity constraints. Using sparse autoencoders (SAEs), we extract feature dictionaries associated with chemically relevant activation patterns. We validate our findings on downstream tasks and find that mechanistic insights can translate to predictive performance in various practical settings.
Industry-Scale Orchestrated Federated Learning for Drug Discovery
Oct 17, 2022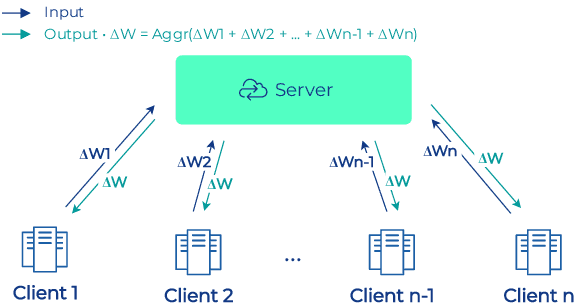
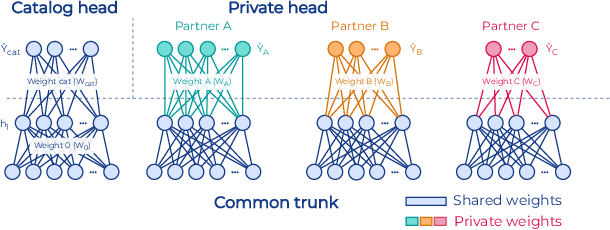
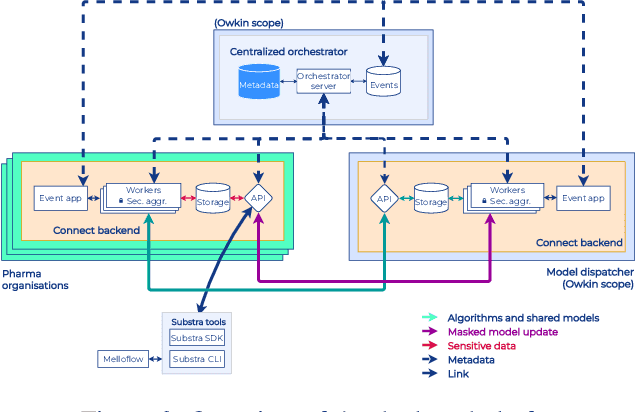
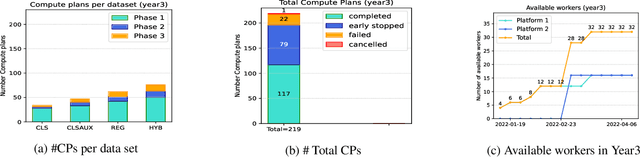
Abstract:To apply federated learning to drug discovery we developed a novel platform in the context of European Innovative Medicines Initiative (IMI) project MELLODDY (grant n{\deg}831472), which was comprised of 10 pharmaceutical companies, academic research labs, large industrial companies and startups. To the best of our knowledge, The MELLODDY platform was the first industry-scale platform to enable the creation of a global federated model for drug discovery without sharing the confidential data sets of the individual partners. The federated model was trained on the platform by aggregating the gradients of all contributing partners in a cryptographic, secure way following each training iteration. The platform was deployed on an Amazon Web Services (AWS) multi-account architecture running Kubernetes clusters in private subnets. Organisationally, the roles of the different partners were codified as different rights and permissions on the platform and administrated in a decentralized way. The MELLODDY platform generated new scientific discoveries which are described in a companion paper.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge